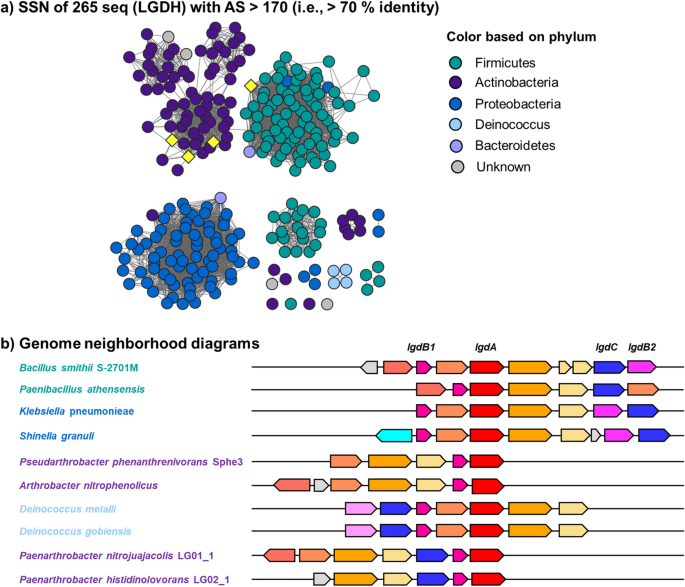
Identification of levoglucosan degradation pathways in bacteria and sequence similarity network analysis
Por um escritor misterioso
Last updated 18 dezembro 2024
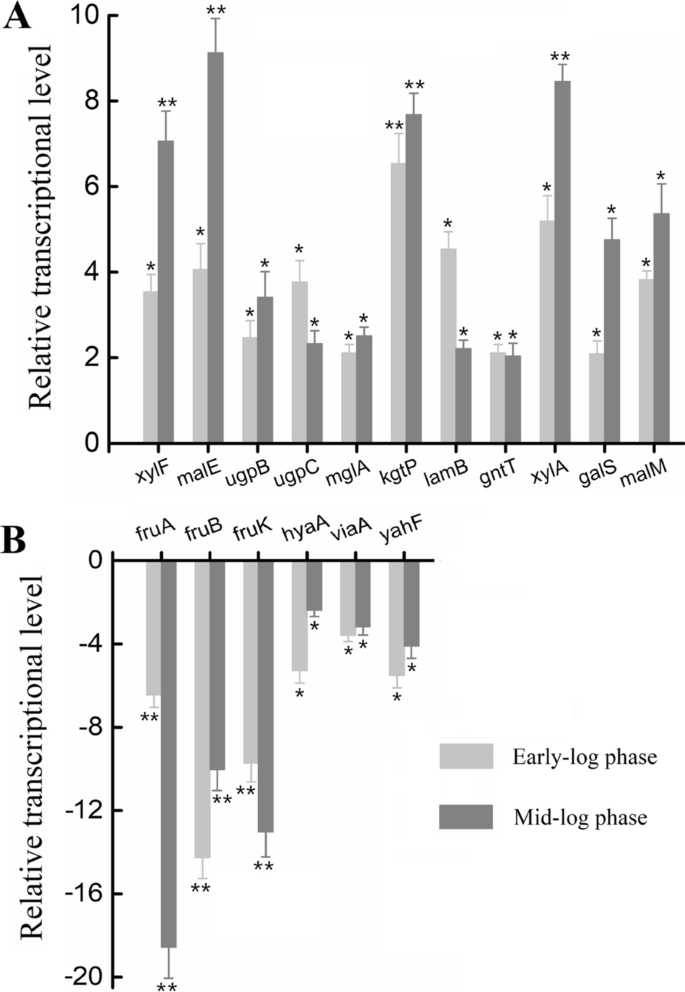
Omics analysis coupled with gene editing revealed potential transporters and regulators related to levoglucosan metabolism efficiency of the engineered Escherichia coli, Biotechnology for Biofuels and Bioproducts

Isolation and Characterization of Levoglucosan-Metabolizing Bacteria

What happens to biomass burning-emitted particles in the ocean? A laboratory experimental approach based on their tracers - ScienceDirect
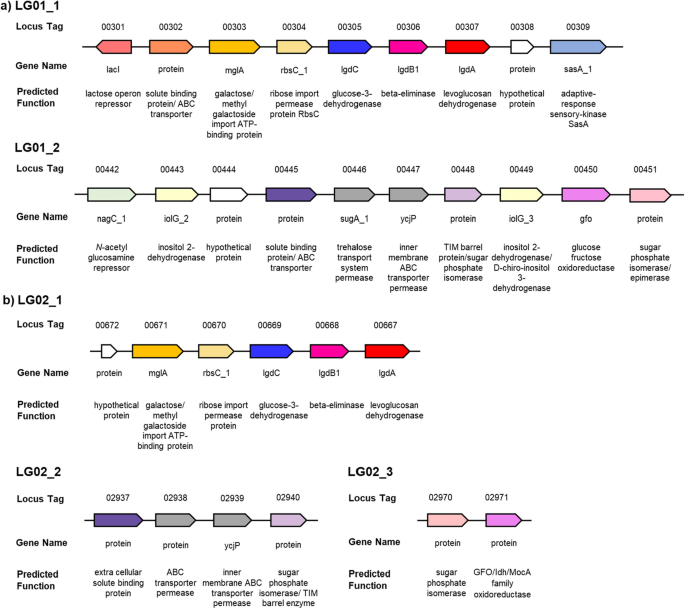
Identification of levoglucosan degradation pathways in bacteria and sequence similarity network analysis
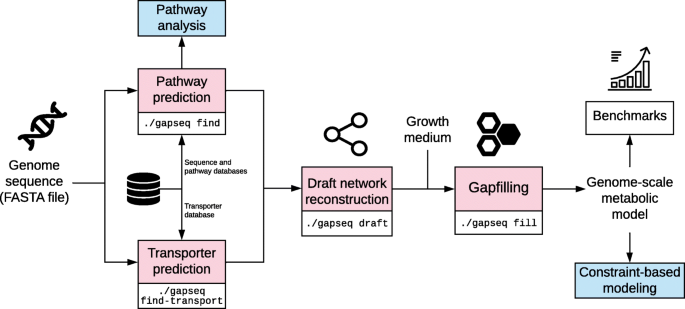
gapseq: informed prediction of bacterial metabolic pathways and reconstruction of accurate metabolic models, Genome Biology

Lipid Production from Dilute Alkali Corn Stover Lignin by Rhodococcus Strains
Archives of Microbiology
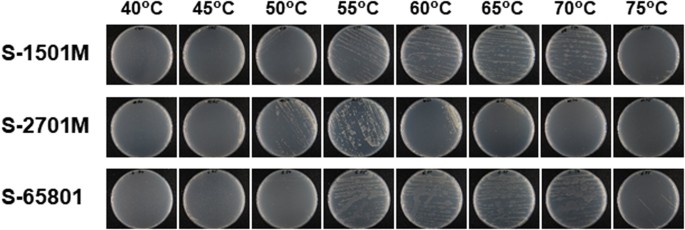
Isolation of levoglucosan-utilizing thermophilic bacteria

Inhibitor tolerance and bioethanol fermentability of levoglucosan-utilizing Escherichia coli were enhanced by overexpression of stress-responsive gene ycfR: The proteomics-guided metabolic engineering - ScienceDirect

Inhibitor tolerance and bioethanol fermentability of levoglucosan-utilizing Escherichia coli were enhanced by overexpression of stress-responsive gene ycfR: The proteomics-guided metabolic engineering - ScienceDirect
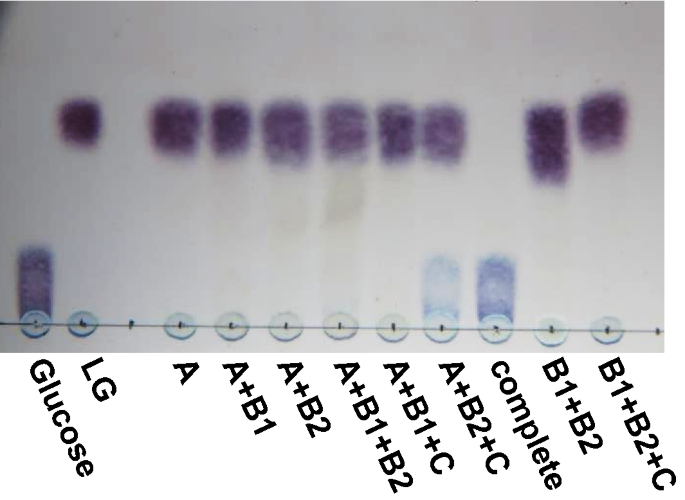
Conversion of levoglucosan into glucose by the coordination of four enzymes through oxidation, elimination, hydration, and reduction
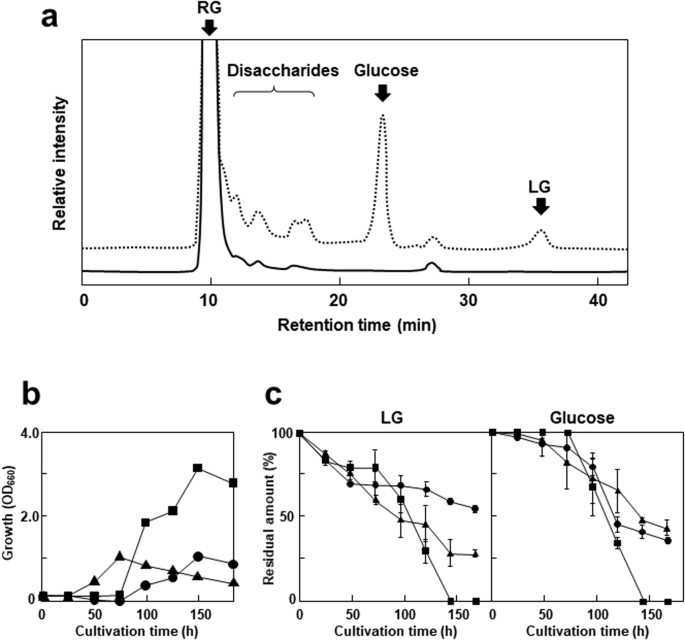
Isolation of levoglucosan-utilizing thermophilic bacteria
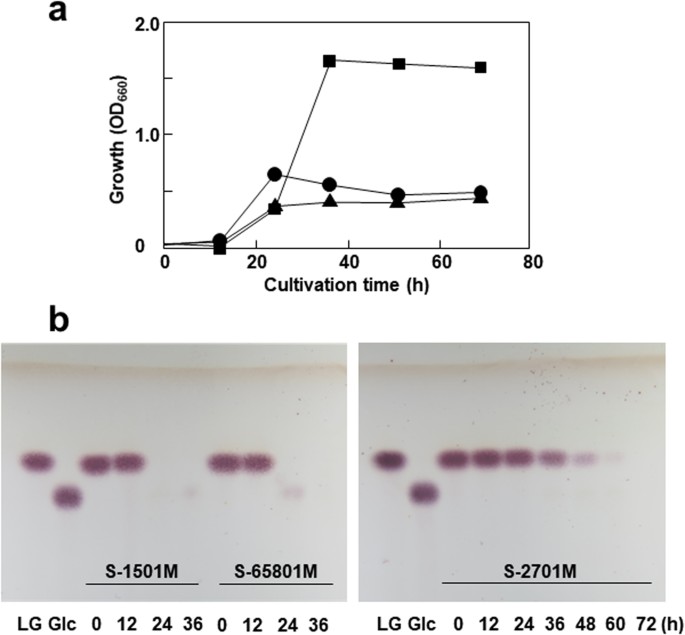
Isolation of levoglucosan-utilizing thermophilic bacteria
Recomendado para você
-
 Stick Nodes Pro - Animator 4.1.3 Free Download18 dezembro 2024
Stick Nodes Pro - Animator 4.1.3 Free Download18 dezembro 2024 -
Stick Nodes Pro - Animator for Android - App Download18 dezembro 2024
-
 Stick Nodes Pro - Animator Mod APK Free Download - FileCR18 dezembro 2024
Stick Nodes Pro - Animator Mod APK Free Download - FileCR18 dezembro 2024 -
 Stick Nodes Pro - Animator IPA Cracked for iOS Free Download18 dezembro 2024
Stick Nodes Pro - Animator IPA Cracked for iOS Free Download18 dezembro 2024 -
 Stick Nodes PC Download Stickman Animation App18 dezembro 2024
Stick Nodes PC Download Stickman Animation App18 dezembro 2024 -
Download free Stick Nodes: Stickman Animator 4.0.6 APK for Android18 dezembro 2024
-
 Stick Nodes (org.fortheloss.sticknodes) 4.1.1 APK 下载 - Android APK - APKsHub18 dezembro 2024
Stick Nodes (org.fortheloss.sticknodes) 4.1.1 APK 下载 - Android APK - APKsHub18 dezembro 2024 -
Stick Nodes: Stickman Animator - Apps on Google Play18 dezembro 2024
-
 Activity, Bolt Animation18 dezembro 2024
Activity, Bolt Animation18 dezembro 2024 -
before and after chaptee 5 animation|TikTok Search18 dezembro 2024
você pode gostar
-
 Puppets for Disability Awareness18 dezembro 2024
Puppets for Disability Awareness18 dezembro 2024 -
 Billfish Fishing18 dezembro 2024
Billfish Fishing18 dezembro 2024 -
 Tim Allen Is Back As Buzz Lightyear In 'Toy Story 5', Tweets, “See Ya Soon, Woody” – Deadline18 dezembro 2024
Tim Allen Is Back As Buzz Lightyear In 'Toy Story 5', Tweets, “See Ya Soon, Woody” – Deadline18 dezembro 2024 -
 Planning Your 2018 Christmas Vacation in Keystone, Colorado - Peak 1 Express18 dezembro 2024
Planning Your 2018 Christmas Vacation in Keystone, Colorado - Peak 1 Express18 dezembro 2024 -
 Garfo Jumbo Tramontina com Lâmina em Aço Inox e Cabo em Madeira Tratada Polywood Vermelho - Garfo Jumbo Tramontina com Lâmina em Aço Inox e Cabo em Madeira Tratada Polywood Vermelho - TRAMONTINA18 dezembro 2024
Garfo Jumbo Tramontina com Lâmina em Aço Inox e Cabo em Madeira Tratada Polywood Vermelho - Garfo Jumbo Tramontina com Lâmina em Aço Inox e Cabo em Madeira Tratada Polywood Vermelho - TRAMONTINA18 dezembro 2024 -
 Create a Blox fruits race awakening v4 Tier List - TierMaker18 dezembro 2024
Create a Blox fruits race awakening v4 Tier List - TierMaker18 dezembro 2024 -
 It's Spooky Month on the Game Boy Color by N-A-R-164 on Newgrounds18 dezembro 2024
It's Spooky Month on the Game Boy Color by N-A-R-164 on Newgrounds18 dezembro 2024 -
 JOGO DE SOBREVIVÊNCIA NO FUNDO DO OCEANO! PERDIDOS NO MAR18 dezembro 2024
JOGO DE SOBREVIVÊNCIA NO FUNDO DO OCEANO! PERDIDOS NO MAR18 dezembro 2024 -
 House of Dragon: Ator sofreu ataques de racismo após ser escolhido para o spin-off de Game of Thrones18 dezembro 2024
House of Dragon: Ator sofreu ataques de racismo após ser escolhido para o spin-off de Game of Thrones18 dezembro 2024 -
 Racing Red 3D Games - Free Car Racing Games To Play Now Online For18 dezembro 2024
Racing Red 3D Games - Free Car Racing Games To Play Now Online For18 dezembro 2024