DREAMS: deep read-level error model for sequencing data applied to low-frequency variant calling and circulating tumor DNA detection, Genome Biology
Por um escritor misterioso
Last updated 18 dezembro 2024
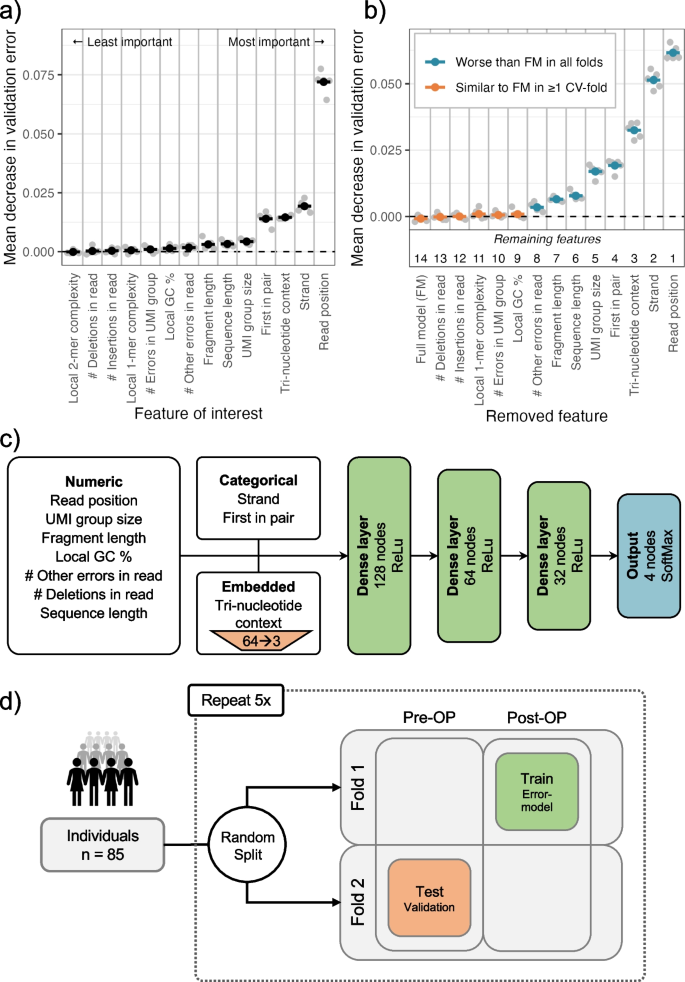
Circulating tumor DNA detection using next-generation sequencing (NGS) data of plasma DNA is promising for cancer identification and characterization. However, the tumor signal in the blood is often low and difficult to distinguish from errors. We present DREAMS (Deep Read-level Modelling of Sequencing-errors) for estimating error rates of individual read positions. Using DREAMS, we develop statistical methods for variant calling (DREAMS-vc) and cancer detection (DREAMS-cc). For evaluation, we generate deep targeted NGS data of matching tumor and plasma DNA from 85 colorectal cancer patients. The DREAMS approach performs better than state-of-the-art methods for variant calling and cancer detection.
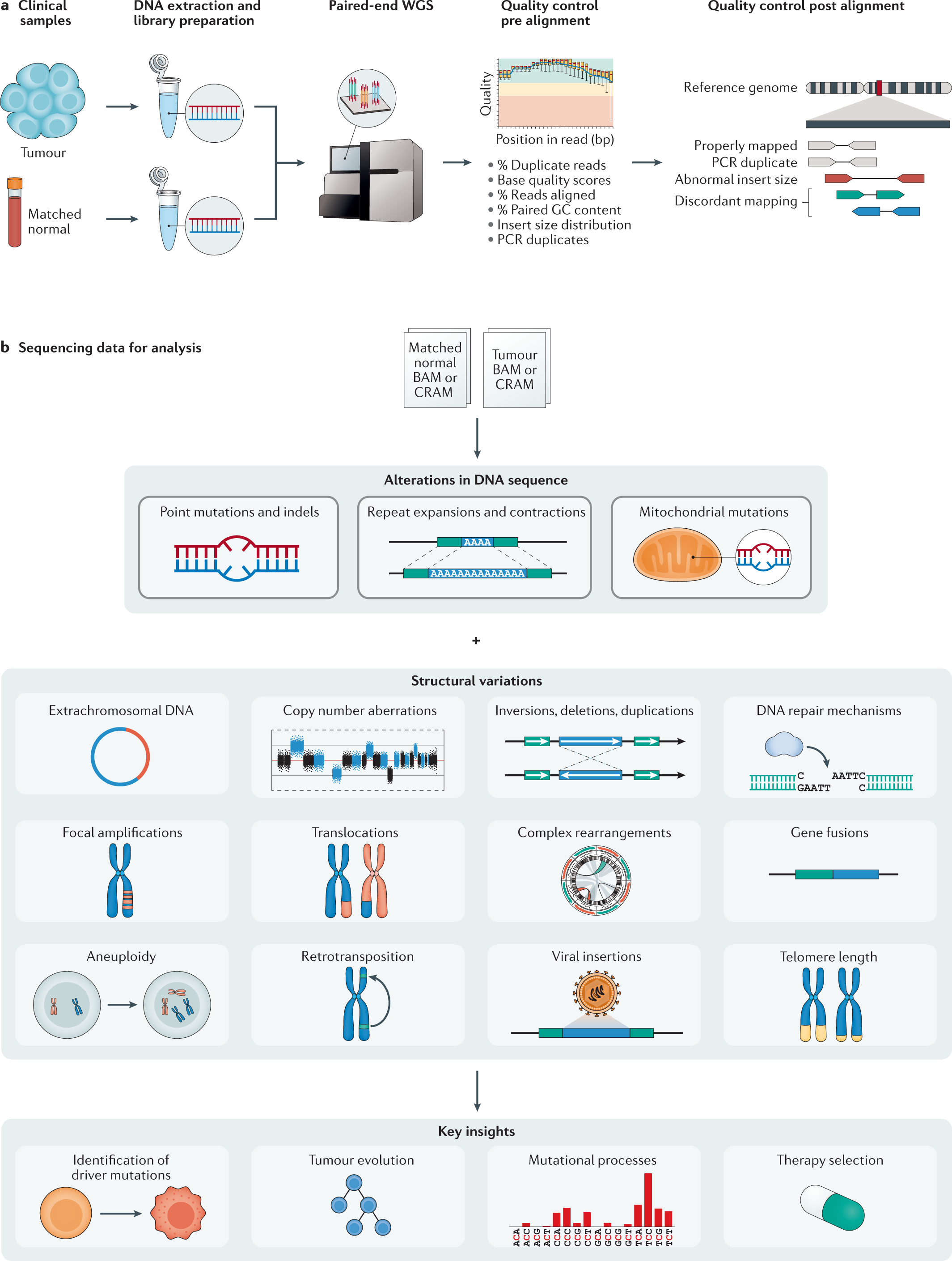
Computational analysis of cancer genome sequencing data

LFMD: detecting low-frequency mutations in high-depth genome

PDF) The changing face of circulating tumor DNA (ctDNA) profiling

Whole genome error-corrected sequencing for sensitive circulating
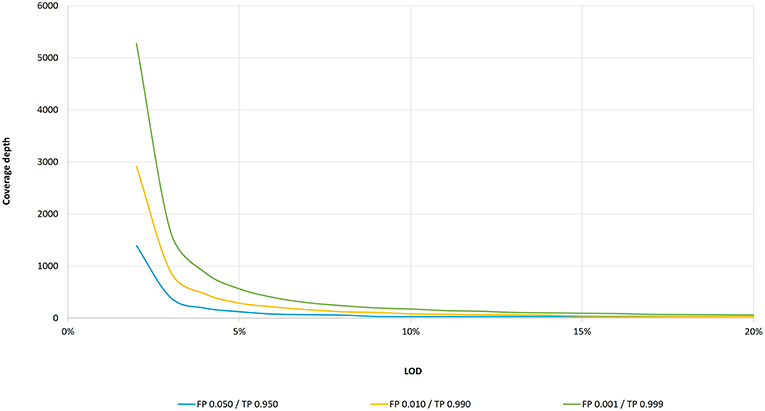
Frontiers Standardization of Sequencing Coverage Depth in NGS

Machine learning guided signal enrichment for ultrasensitive
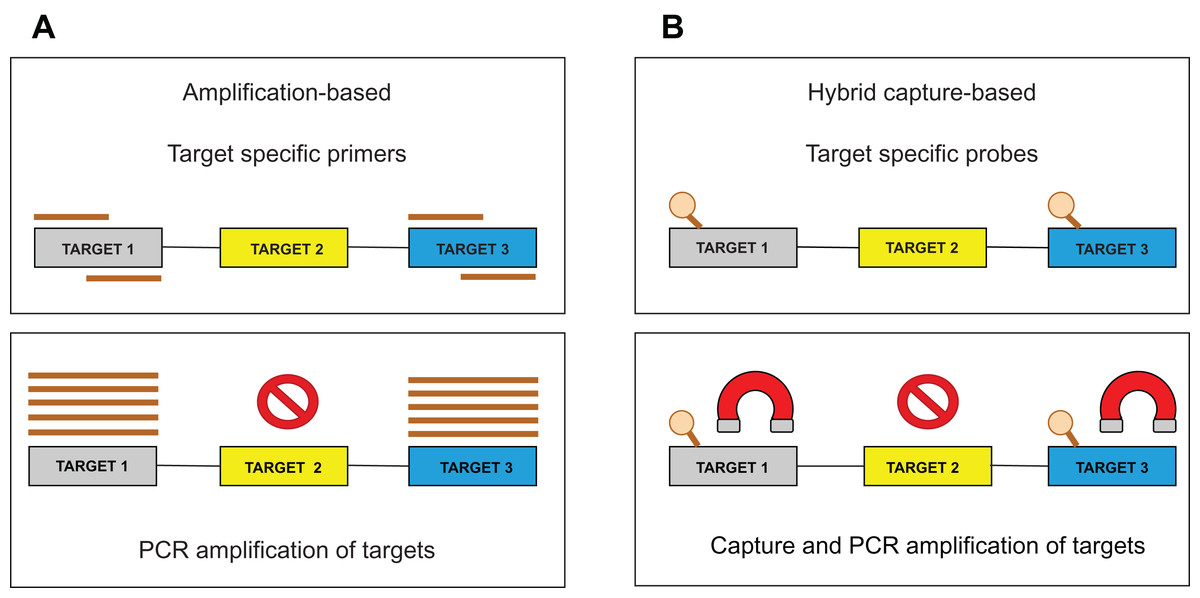
Bioinformatic strategies for the analysis of genomic aberrations

PDF) DREAMS: deep read-level error model for sequencing data

Machine learning guided signal enrichment for ultrasensitive

Analytical and Clinical Validation of an Amplicon-based Next

Integration of intra-sample contextual error modeling for improved
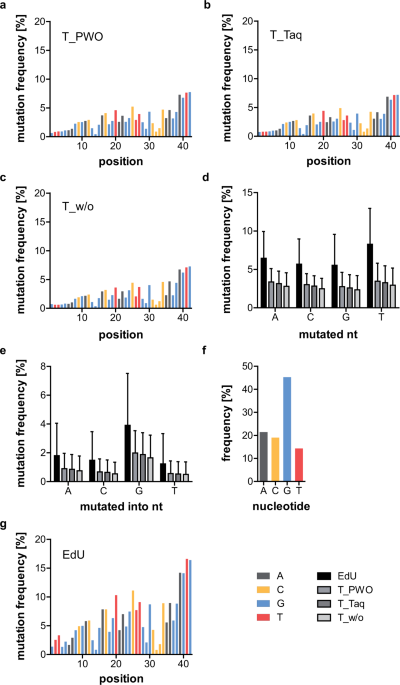
Systematic evaluation of error rates and causes in short samples
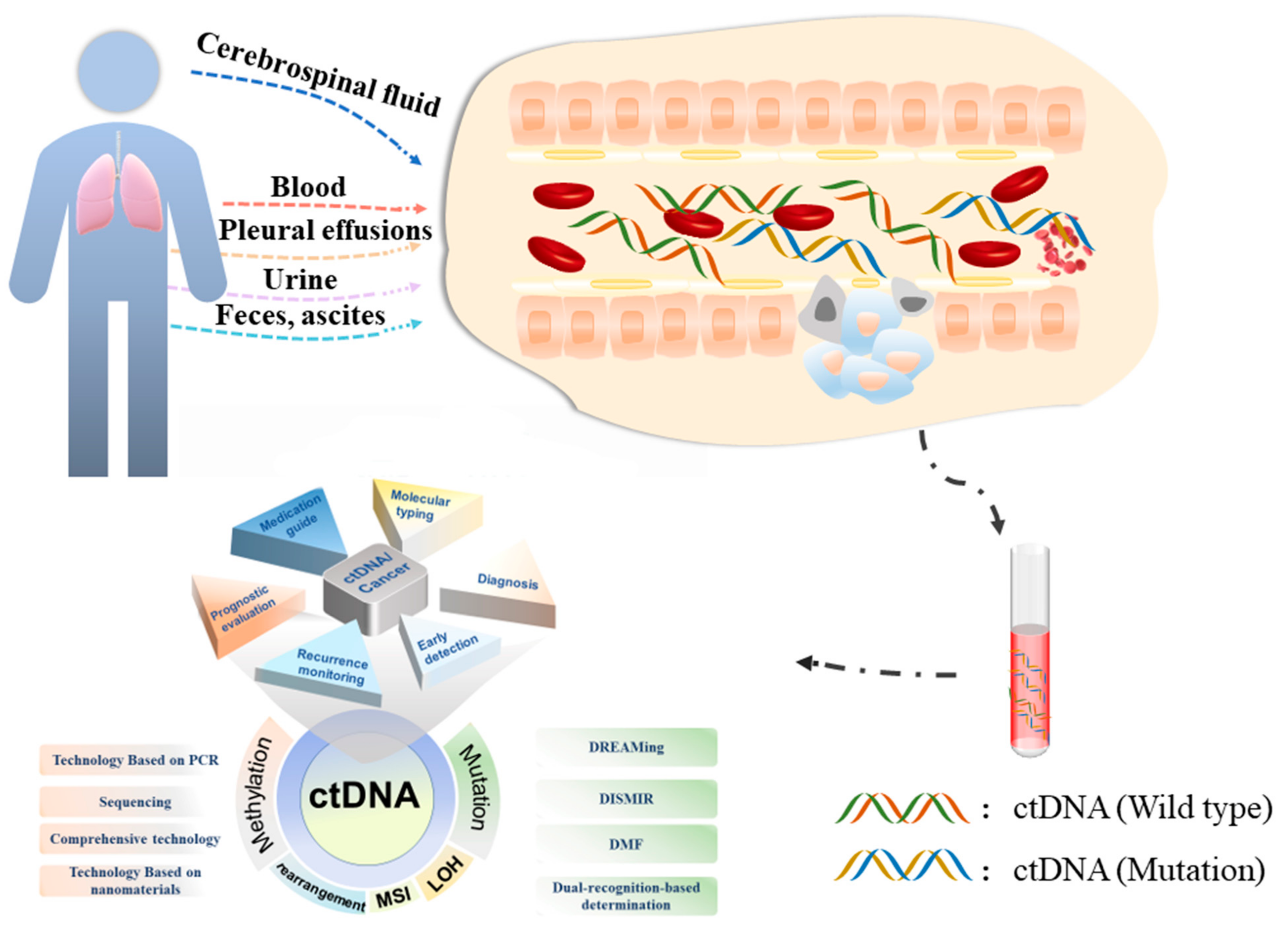
Cancers, Free Full-Text
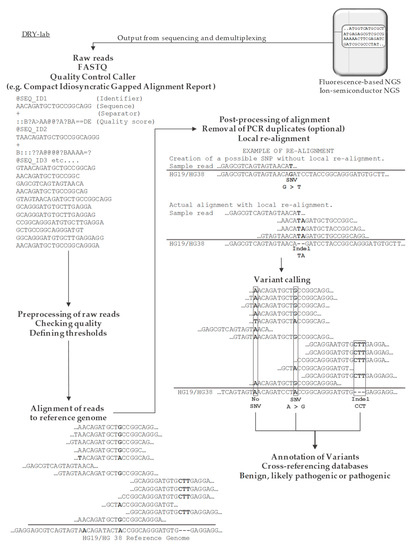
Cancers, Free Full-Text
Recomendado para você
-
 The SCP Foundation Joke Series, List of Deaths Wiki18 dezembro 2024
The SCP Foundation Joke Series, List of Deaths Wiki18 dezembro 2024 -
 Don't forget about SCP 7143-J : r/DankMemesFromSite1918 dezembro 2024
Don't forget about SCP 7143-J : r/DankMemesFromSite1918 dezembro 2024 -
 OC snubbed from an /x/ thread, SCP Foundation18 dezembro 2024
OC snubbed from an /x/ thread, SCP Foundation18 dezembro 2024 -
 press F for 7143-J : r/DankMemesFromSite1918 dezembro 2024
press F for 7143-J : r/DankMemesFromSite1918 dezembro 2024 -
open_science_fellowship_project/resources/example_data/derivatives/freesurfer/sub-01/scripts/recon-all.local-copy at master · PeerHerholz/open_science_fellowship_project · GitHub18 dezembro 2024
-
 Machimaho: I Messed Up and Made the Wrong Person Into a Magical Girl! - AnimeSuki Forum18 dezembro 2024
Machimaho: I Messed Up and Made the Wrong Person Into a Magical Girl! - AnimeSuki Forum18 dezembro 2024 -
Miscellaneous SCPs, SCP Meta Wiki18 dezembro 2024
-
Gorefield - Parte 2 [SCP-3166] #gorefield #garfield #scp3166 #scp #cr18 dezembro 2024
-
 Journal of Comparative Neurology, Systems Neuroscience Journal18 dezembro 2024
Journal of Comparative Neurology, Systems Neuroscience Journal18 dezembro 2024 -
 Solubility Enhancement of Hydrophobic Substances in Water/Cyrene Mixtures: A Computational Study18 dezembro 2024
Solubility Enhancement of Hydrophobic Substances in Water/Cyrene Mixtures: A Computational Study18 dezembro 2024
você pode gostar
-
How to Get The Lake Trio in Pixelmon 2021 #minecraft #uxie #azelf18 dezembro 2024
-
 Immortals of Aveum - Kalthus, All Golden Chests & Shroudfanes18 dezembro 2024
Immortals of Aveum - Kalthus, All Golden Chests & Shroudfanes18 dezembro 2024 -
 Elden Ring: walkthrough of Fort Faroth to obtain Radagon's18 dezembro 2024
Elden Ring: walkthrough of Fort Faroth to obtain Radagon's18 dezembro 2024 -
Meaning of So American by Portugal. The Man18 dezembro 2024
-
 The Best Minecraft Mods 1.19 - Minecraft Guide - IGN18 dezembro 2024
The Best Minecraft Mods 1.19 - Minecraft Guide - IGN18 dezembro 2024 -
 Como conseguir o emoji do fino senhores/moai parte 218 dezembro 2024
Como conseguir o emoji do fino senhores/moai parte 218 dezembro 2024 -
 Wonder Woman vs. Silver Swan BLOODLINES clip18 dezembro 2024
Wonder Woman vs. Silver Swan BLOODLINES clip18 dezembro 2024 -
 Good Girl Hair Mist 30ml - Carolina Herrera - Condessa Cosméticos e Perfumaria18 dezembro 2024
Good Girl Hair Mist 30ml - Carolina Herrera - Condessa Cosméticos e Perfumaria18 dezembro 2024 -
 Realeza Absoluta - Pokemon - Epic Game18 dezembro 2024
Realeza Absoluta - Pokemon - Epic Game18 dezembro 2024 -
Colar Fairy Tail Anime Mangá Cosplay Pingente Guilda18 dezembro 2024
![Gorefield - Parte 2 [SCP-3166] #gorefield #garfield #scp3166 #scp #cr](https://www.tiktok.com/api/img/?itemId=7226380769608862982&location=0&aid=1988)

